Functional magnetic resonance imaging (fMRI) has transformed our understanding of the human brain, enabling researchers to map cognitive functions to specific neural structures with millimeter precision. Yet the field faces a reproducibility challenge: a landmark 2022 study in Nature demonstrated that brain-wide association studies require thousands of participants to achieve reliable, replicable results [1]. Much of this variability stems not from biology but from analytical choices.
The Reproducibility Problem in Neuroimaging
Every fMRI analysis involves dozens of methodological decisions: preprocessing parameters, motion correction thresholds, statistical models, multiple comparison corrections, and visualization choices. A 2022 study found that the choice of processing pipeline alone drives considerable variability in the location of identified brain markers [2]. Different laboratories analyzing identical data can reach different conclusions simply due to pipeline selection.
This "researcher degrees of freedom" problem compounds when analyses lack complete documentation. Published methods sections often omit critical parameters, software versions, or preprocessing steps that would enable exact replication. The result is a literature where effect sizes are inflated, false positives accumulate, and genuine findings remain difficult to distinguish from analytical artifacts.
The General Linear Model: Foundation of Task fMRI
Most task-based fMRI analyses rely on the General Linear Model (GLM), a statistical framework that models the expected blood-oxygen-level-dependent (BOLD) signal as a linear combination of experimental conditions convolved with the hemodynamic response function.
The GLM approach involves several key steps:
- Design matrix construction: Encoding experimental conditions and their timing
- Model fitting: Estimating beta coefficients for each voxel
- Statistical inference: Computing t-statistics and applying significance thresholds
- Cluster correction: Controlling family-wise error rates across thousands of voxels
Each step requires parameter choices that affect results. The canonical p < 0.001 threshold with cluster-extent correction has become standard, but alternatives exist and may be more appropriate depending on study goals.
Anatomy of a Reproducible Analysis
A truly reproducible neuroimaging analysis requires complete specification of:
Input data: File formats, coordinate systems, acquisition parameters, and data quality metrics. The Brain Imaging Data Structure (BIDS) standard provides a framework for organizing neuroimaging data consistently.
Processing environment: Exact software versions, dependencies, and computational environment. Without version control, analyses may produce different results when re-run with updated libraries.
Analysis parameters: Every threshold, smoothing kernel, and model specification must be documented.
Output validation: Statistical maps, cluster summaries, and quality assurance metrics that enable verification of results.
The following example demonstrates what complete pipeline documentation looks like for a task-based fMRI GLM analysis:
Input Specification
Figure 1: Multi-slice brain activity captured via fMRI. These axial slices serve as input to the automated analysis pipeline, which performs task-aligned GLM analysis to detect regions showing statistically significant activation.
A well-specified input includes the fMRI volume dimensions (e.g., 20x20x10 voxels), number of timepoints (e.g., 100 TRs), and accompanying event files that encode experimental timing.
Activation Detection Results
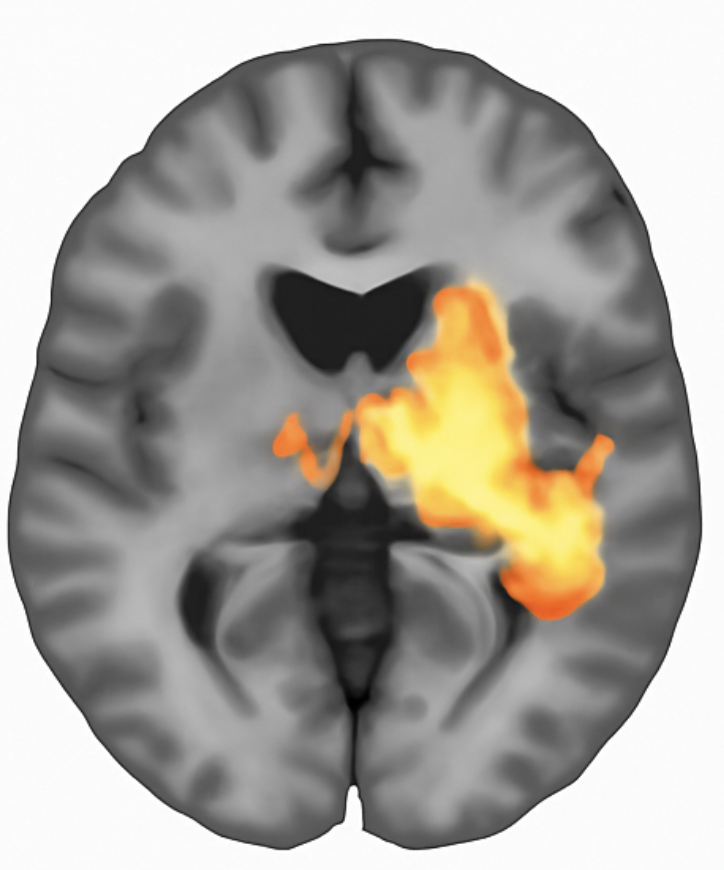
Figure 2: Activation map generated by voxelwise GLM analysis. Colored regions indicate voxels exceeding the statistical threshold (p < 0.001, uncorrected). The analysis identified 11 significant voxels representing 0.275% of the brain volume, with peak activation at coordinates [18, 16, 3] achieving t = 3.89.
The GLM output provides interpretable metrics: the number and percentage of significant voxels, cluster locations and sizes, and peak statistics with precise coordinates enabling cross-study comparison.
Structured Output Documentation
Complete reproducibility requires machine-readable output specifications:
{
"analysis_id": "glm_task_v1",
"model": {
"type": "GLM",
"design_matrix_shape": [100, 2],
"regressors": ["task", "intercept"],
"threshold_p": 0.001
},
"results": {
"significant_voxels": 11,
"significance_fraction": 0.00275,
"peak_activation": {
"coordinates": [18, 16, 3],
"t_value": 3.89,
"p_value": 0.0004
},
"cluster_summary": [
{"cluster_id": 1, "size": 6, "peak_t": 3.89},
{"cluster_id": 2, "size": 3, "peak_t": 3.45}
]
}
}Figure 3: Structured analysis results in JSON format. Machine-readable outputs enable automated meta-analyses, quality assurance checks, and exact replication.
The Automation Advantage
Manually documenting every parameter, version, and output is tedious and error-prone. AI agents configured for neuroimaging can automate this process:
Hypothesis translation: Convert natural language research questions into formal statistical models with appropriate design matrices.
Pipeline generation: Construct complete analysis workflows with BIDS-compliant inputs, standardized preprocessing, and validated statistical methods.
Automatic documentation: Generate comprehensive manifests capturing every analytical decision, software version, and intermediate output.
Quality assurance: Flag potential issues such as excessive motion, registration failures, or statistical anomalies before they propagate to conclusions.
From Hours to Minutes
Traditional fMRI analysis requires iterating through preprocessing, model specification, statistical testing, and visualization, often over days or weeks. Each iteration risks introducing undocumented changes. Automated pipelines compress this timeline while enforcing documentation standards.
More importantly, automation separates the creative work of hypothesis generation from the mechanical work of implementation. Researchers can focus on biological questions while agents handle the translation to reproducible code.
Toward Cumulative Science
The neuroimaging field has recognized reproducibility as essential. Large-scale initiatives like the Human Connectome Project and UK Biobank provide standardized datasets. The BIDS standard enforces data organization. Pre-registration reduces analytical flexibility.
Automated analysis pipelines represent the next step: ensuring that every published finding comes with complete, executable documentation. When any researcher can reproduce any analysis exactly, science becomes truly cumulative.
References
[1] Marek S, et al. "Reproducible brain-wide association studies require thousands of individuals." Nature. 2022;603(7902):654-660. https://doi.org/10.1038/s41586-022-04492-9
[2] Zhou X, et al. "Choice of Voxel-based Morphometry processing pipeline drives variability in the location of neuroanatomical brain markers." Communications Biology. 2022;5(1):913. https://doi.org/10.1038/s42003-022-03880-1
[3] Klapwijk ET, et al. "Opportunities for increased reproducibility and replicability of developmental neuroimaging." Developmental Cognitive Neuroscience. 2021;47:100902. https://doi.org/10.1016/j.dcn.2020.100902
[4] Rosenblatt M, et al. "Data leakage inflates prediction performance in connectome-based machine learning models." Nature Communications. 2024;15(1):1829. https://doi.org/10.1038/s41467-024-46150-w

